Estimating the Effective Reproduction Number of COVID-19 in Canada
Louis Rossouw
2023-02-24 21:35:11 EST`
1 Introduction
This paper contains estimates for the effective reproduction number
\(R_{t,m}\) over time \(t\) in various provinces (and health
regions) \(m\) of Canada. This is done
using the methodology as described in [1].
These have been implemented in R using EpiEstim package
[2] which is what is used here. The
methodology and assumptions are described in more detail here.
This paper and it’s results should be updated roughly daily and is available online.
2 Updates
As this paper is updated over time this section will summarise significant changes. The code producing this paper is tracked using Git. The Git commit hash for this project at the time of generating this paper was 6cab0e089bd06c9688728ab585072be9bce67169.
The following major updates have been made:
- On 29 May 2021 all plots and maps were updated to consistently not plot reproduction number estimates where the 95% confidence interval associated with that estimate is wider than 1.
3 Data
Data is downloaded from the Git repository associated with [3]. This contains the daily cases and deaths reported for Canada by province and health regions.
Repatriated cases are removed and provinces as found in [4] are grouped per the table below:
| Province | Province Grouping |
|---|---|
| Alberta | Alberta |
| British Columbia | British Columbia |
| Manitoba | Manitoba |
| New Brunswick | Maritime Provinces |
| Newfoundland and Labrador | Maritime Provinces |
| Nova Scotia | Maritime Provinces |
| Nunavut | Territories |
| Northwest Territories | Territories |
| Ontario | Ontario |
| Prince Edward Island | Maritime Provinces |
| Quebec | Quebec |
| Saskatchewan | Saskatchewan |
| Yukon | Territories |
The only adjustments to the data relates to large numbers of deaths reported in Ontario on 2 and 3 October 2020 “due to a data review”. 111 of the deaths reported on these two dates are deaths that occurred during the prior spring and summer (see [5] and [6]). Based on this these deaths were removed from 2 and 3 October and added back in prior days in proportion to deaths reported up un till 30 September 2020. The net effect is no change in reported deaths, but a peak in October is avoided which would have biased estimates.
4 Methodology
The methodology is described in detail here.
5 Results
5.1 Cases
Below a 7-day moving average daily case count is plotted by province on a log scale:
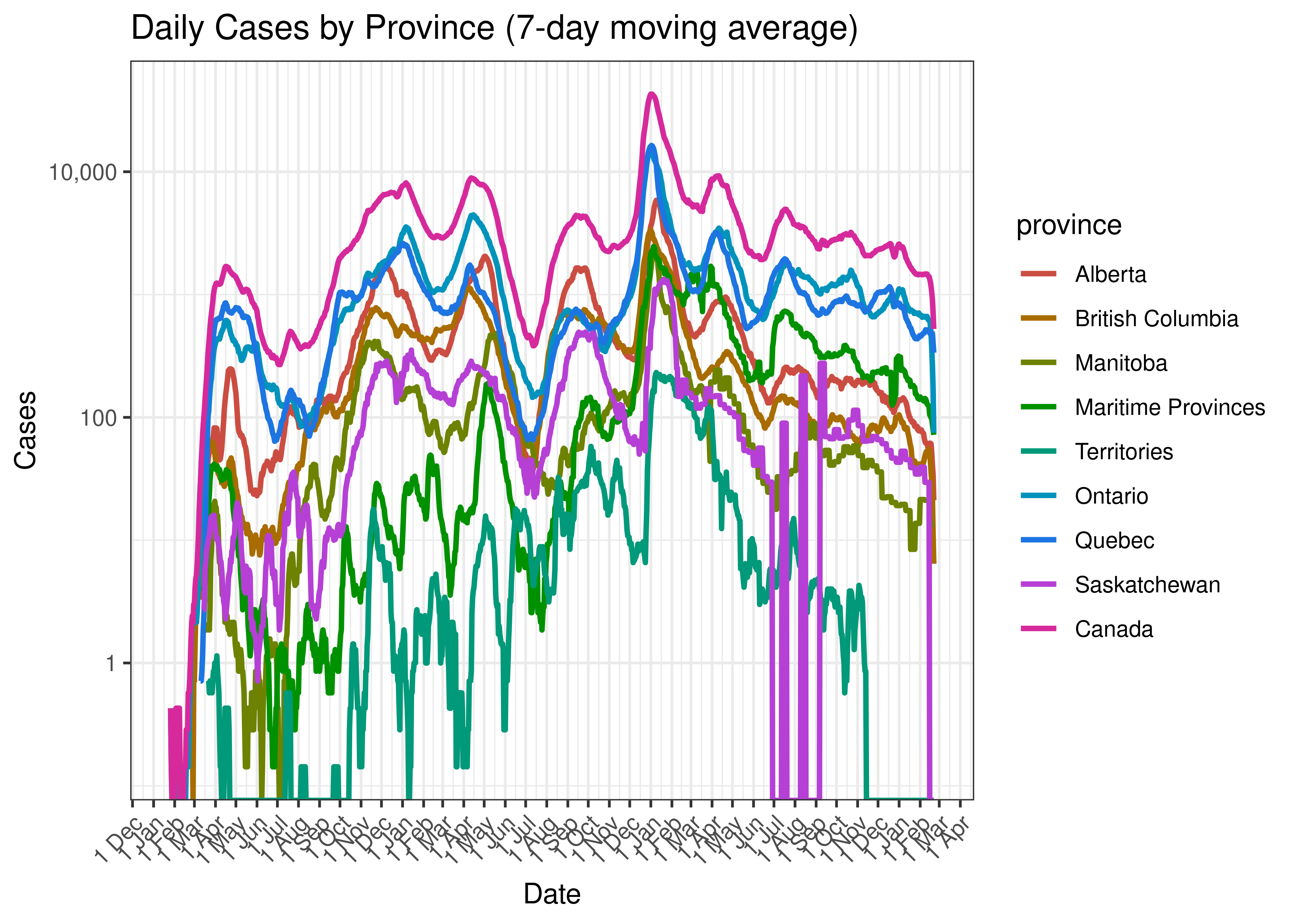
Daily Cases by Province (7-day moving average)
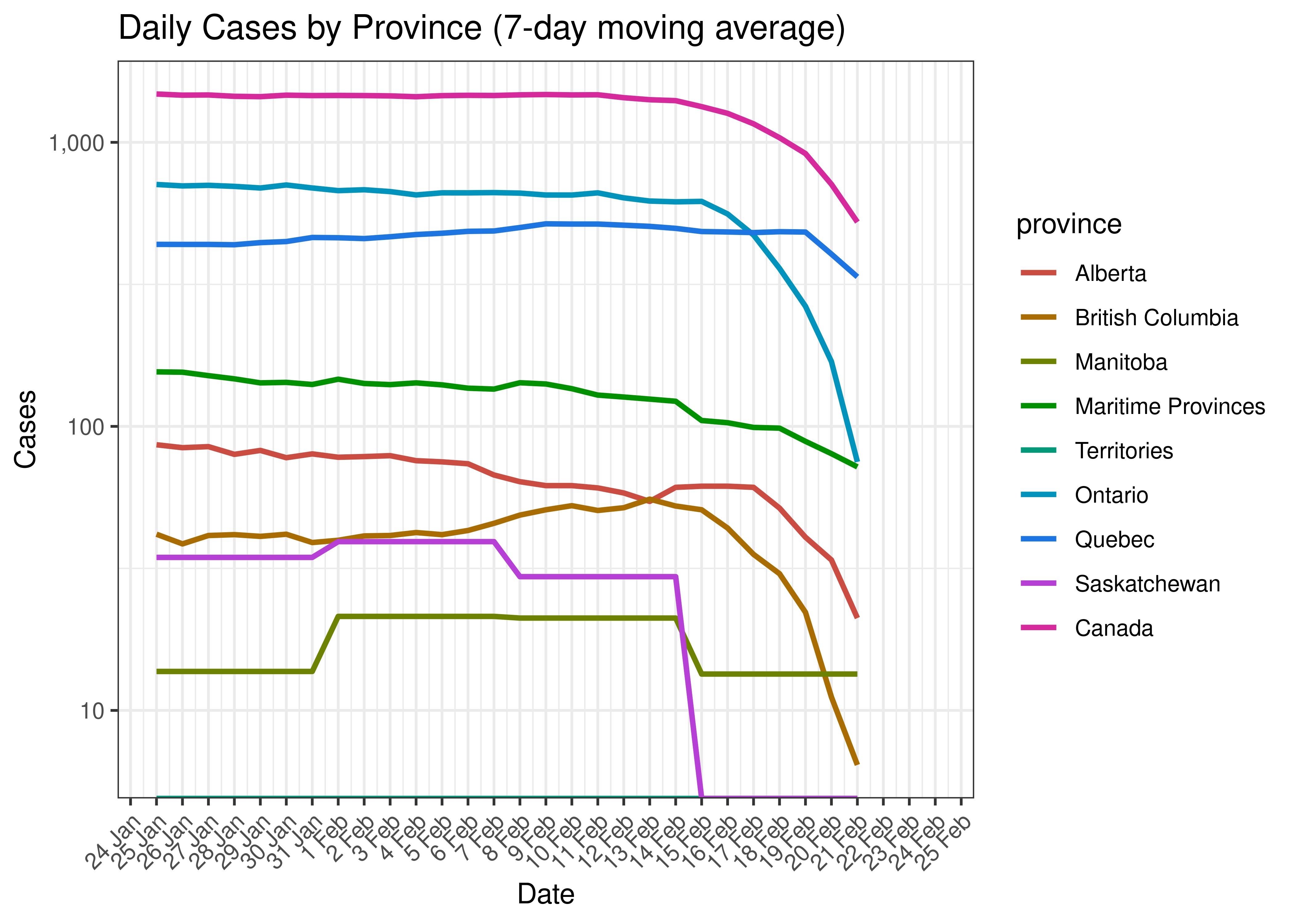
Daily Cases by Province for Last 30 Days (7-day moving average)
5.2 Deaths
Below a 14-day moving average of daily reported deaths by province is plotted on a log scale.
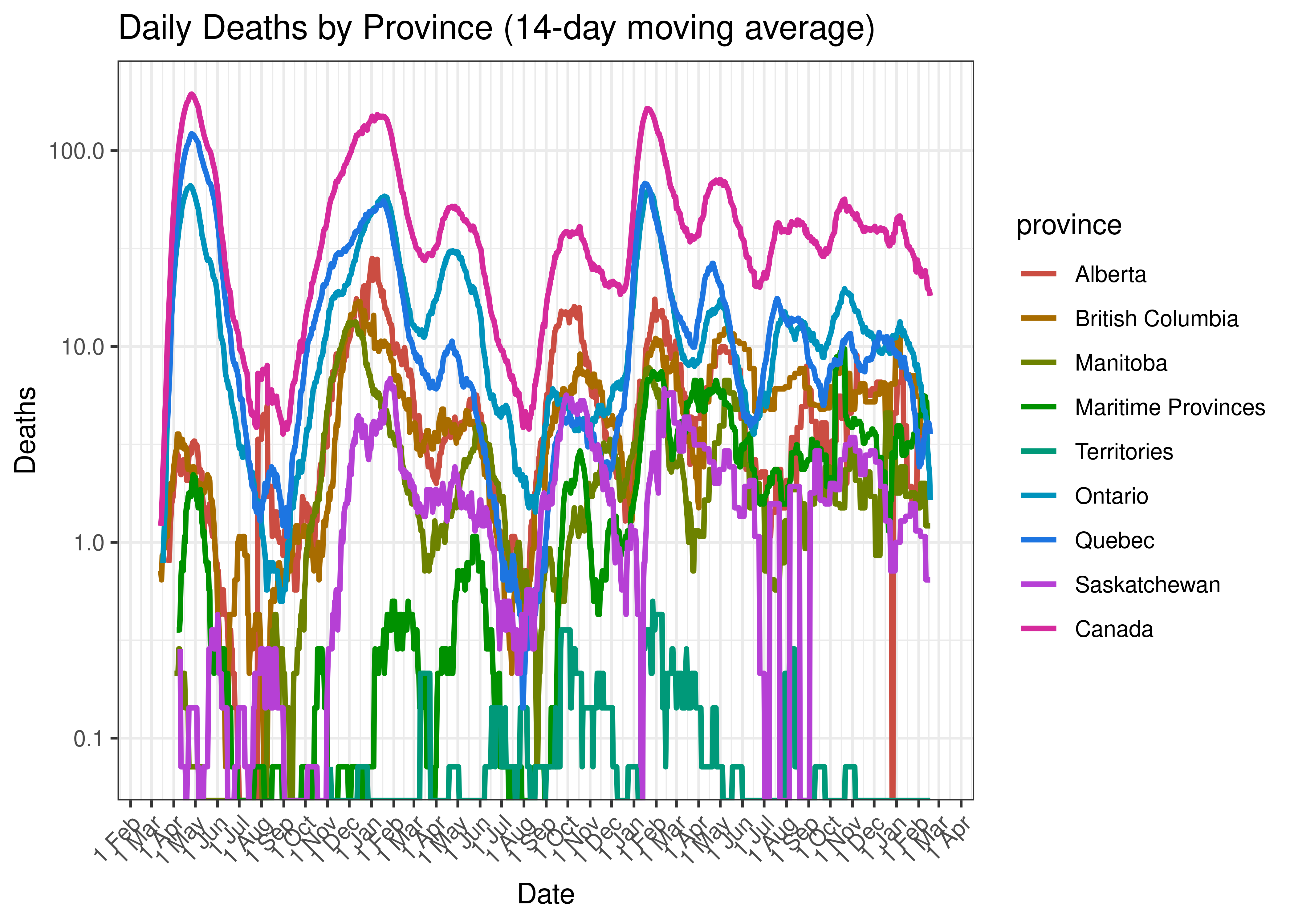
Daily Deaths by Province (14-day moving average)
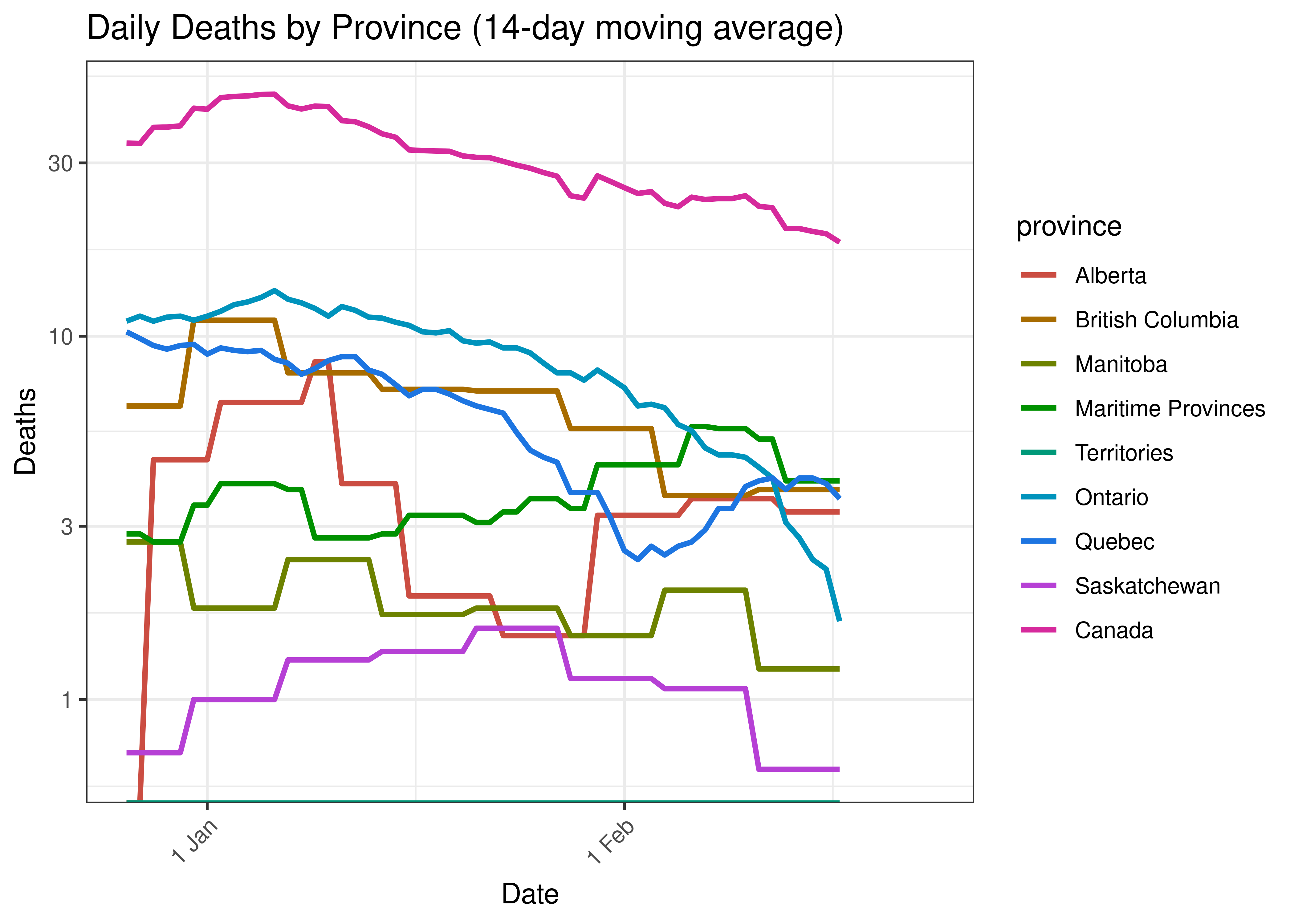
Daily Deaths by Province for Last 60 Days (14-day moving average)
5.3 Current reproduction number estimates by Province
Below current (last weekly) reproduction number estimates are tabulated.
| province | Estimated Type | Daily Count (Last Week) | Week Ending | Reproduction Number [95% Confidence Interval] |
|---|---|---|---|---|
| Alberta | cases | 21 | 2023-02-24 | 0.37 [0.31 - 0.44] |
| Alberta | deaths | 3 | 2023-02-24 | 0.89 [0.55 - 1.32] |
| British Columbia | cases | 6 | 2023-02-24 | 0.16 [0.11 - 0.22] |
| British Columbia | deaths | 5 | 2023-02-24 | 1.15 [0.80 - 1.56] |
| Manitoba | cases | 13 | 2023-02-24 | 0.89 [0.72 - 1.09] |
| Manitoba | deaths | 0 | 2023-02-24 | 0.27 [0.01 - 1.09] |
| Maritime Provinces | cases | 72 | 2023-02-24 | 0.68 [0.62 - 0.75] |
| Maritime Provinces | deaths | 2 | 2023-02-24 | 0.47 [0.25 - 0.78] |
| Territories | cases | 0 | 2023-02-24 | 5.05 [0.12 - 18.73] |
| Territories | deaths | 0 | 2023-02-24 | 5.00 [0.12 - 18.42] |
| Ontario | cases | 75 | 2023-02-24 | 0.15 [0.13 - 0.17] |
| Ontario | deaths | 0 | 2023-02-24 | 0.12 [0.01 - 0.33] |
| Quebec | cases | 336 | 2023-02-24 | 0.69 [0.66 - 0.72] |
| Quebec | deaths | 2 | 2023-02-24 | 0.49 [0.28 - 0.77] |
| Saskatchewan | cases | 0 | 2023-02-24 | 0.02 [0.00 - 0.10] |
| Saskatchewan | deaths | 0 | 2023-02-24 | 0.49 [0.01 - 1.95] |
| Canada | cases | 524 | 2023-02-24 | 0.43 [0.40 - 0.46] |
| Canada | deaths | 12 | 2023-02-24 | 0.61 [0.48 - 0.75] |
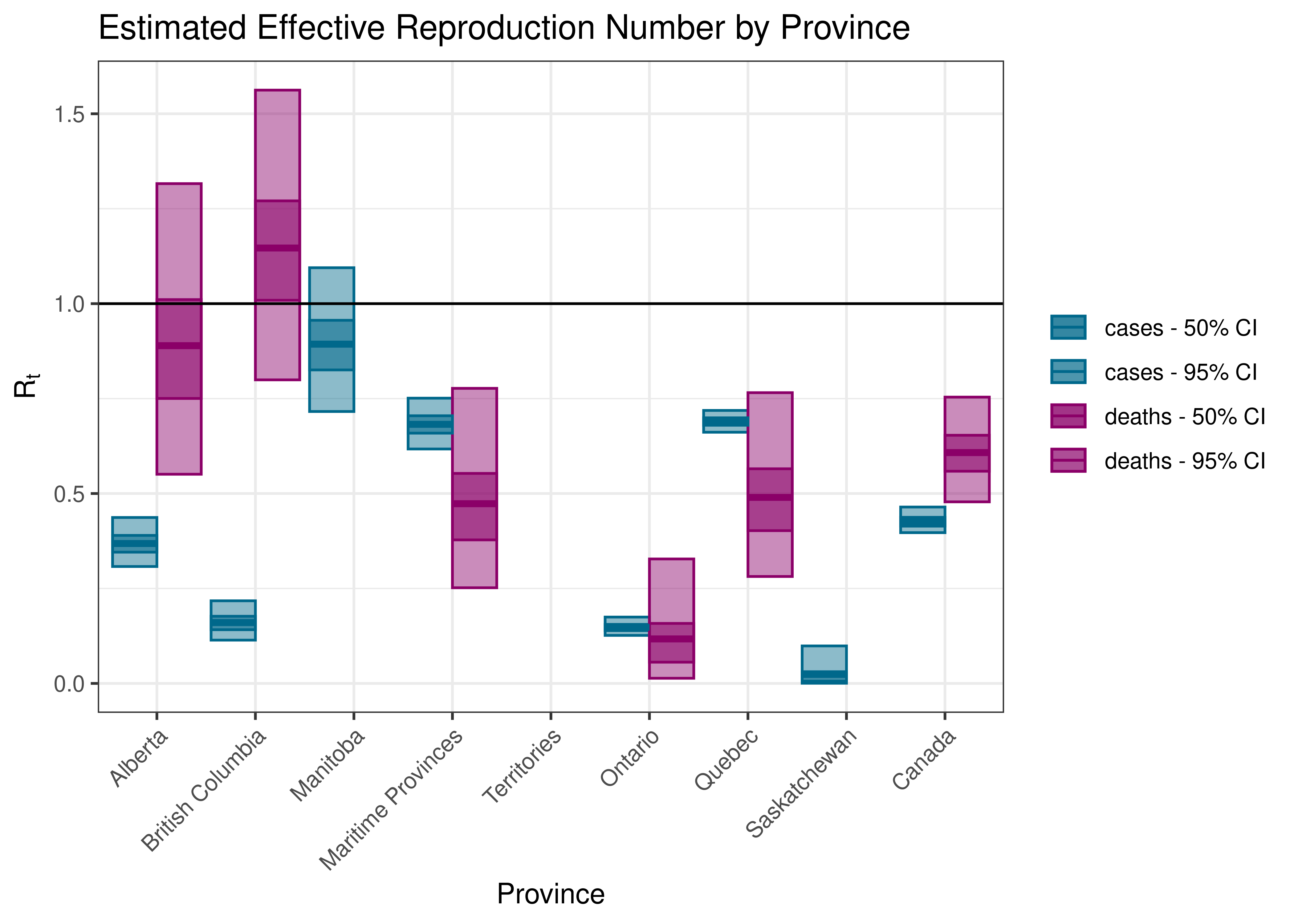
Estimated Effective Reproduction Number by Province
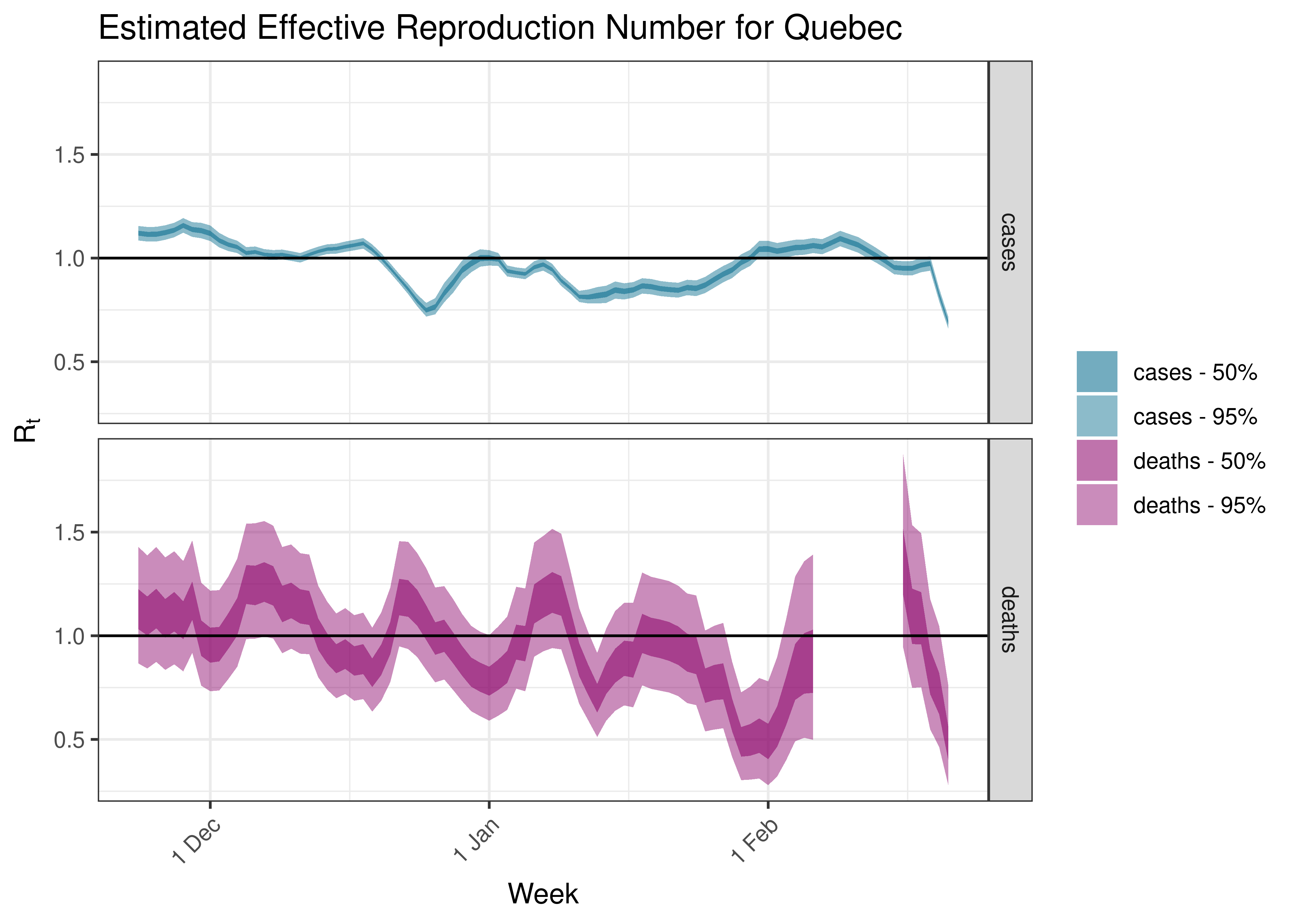
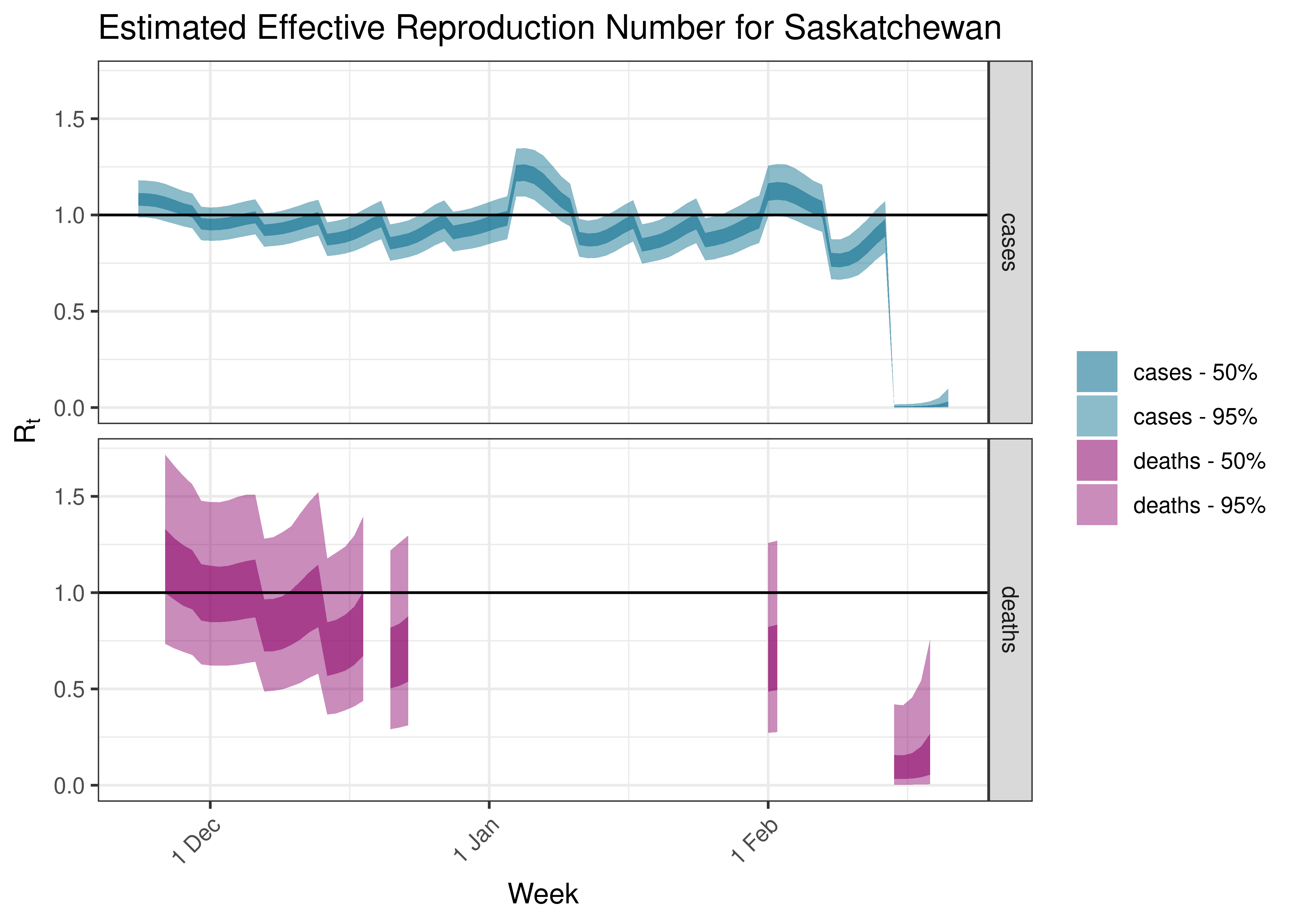
5.4 Estimated Effective Reproduction Number for Canada over Time
Below we plot results for Canada as a whole.
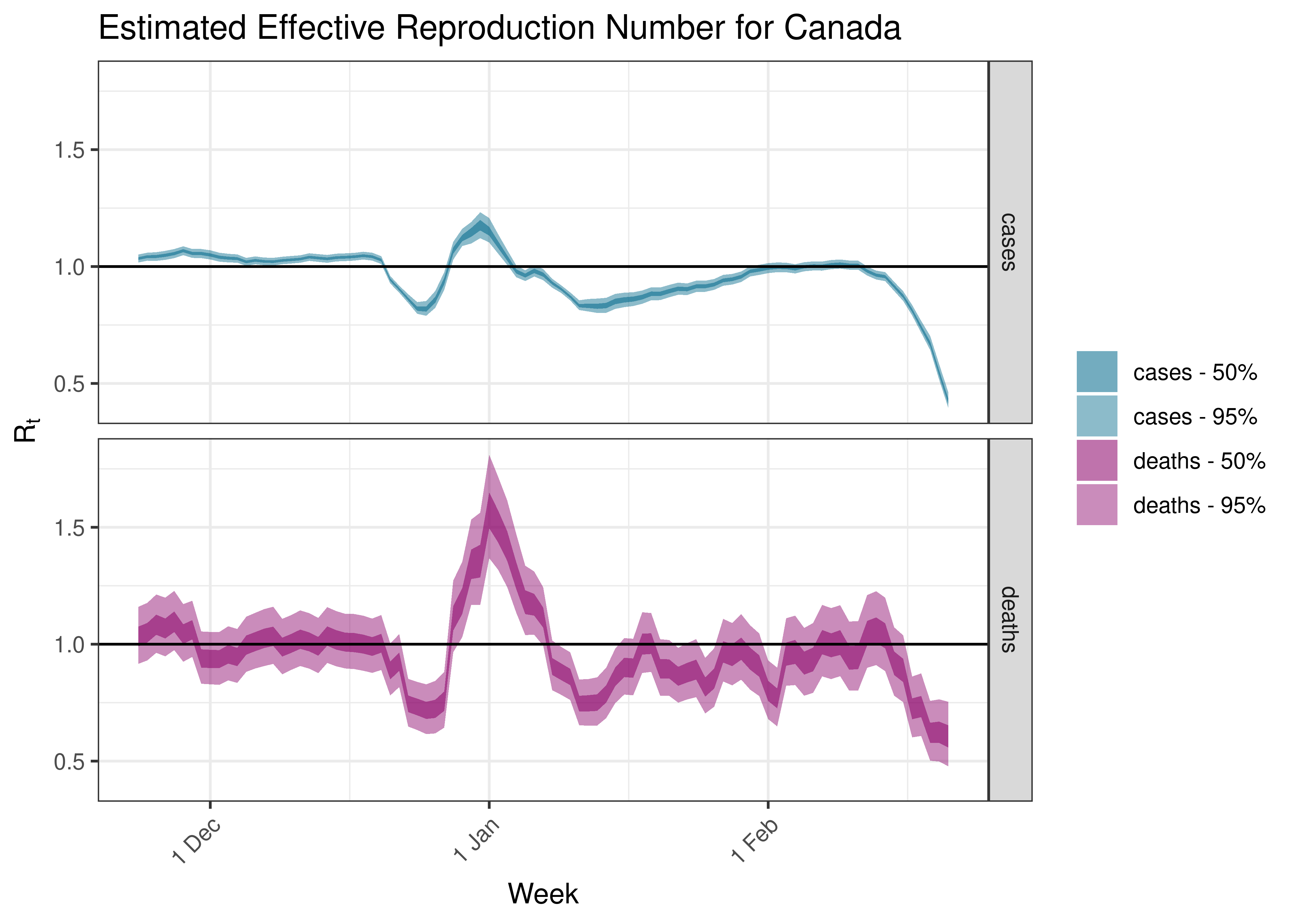
Estimated Effective Reproduction Number for Canada over Time
5.5 Estimated Effective Reproduction Number for Provinces over Time
Below we plot results for each province.
5.5.1 Alberta
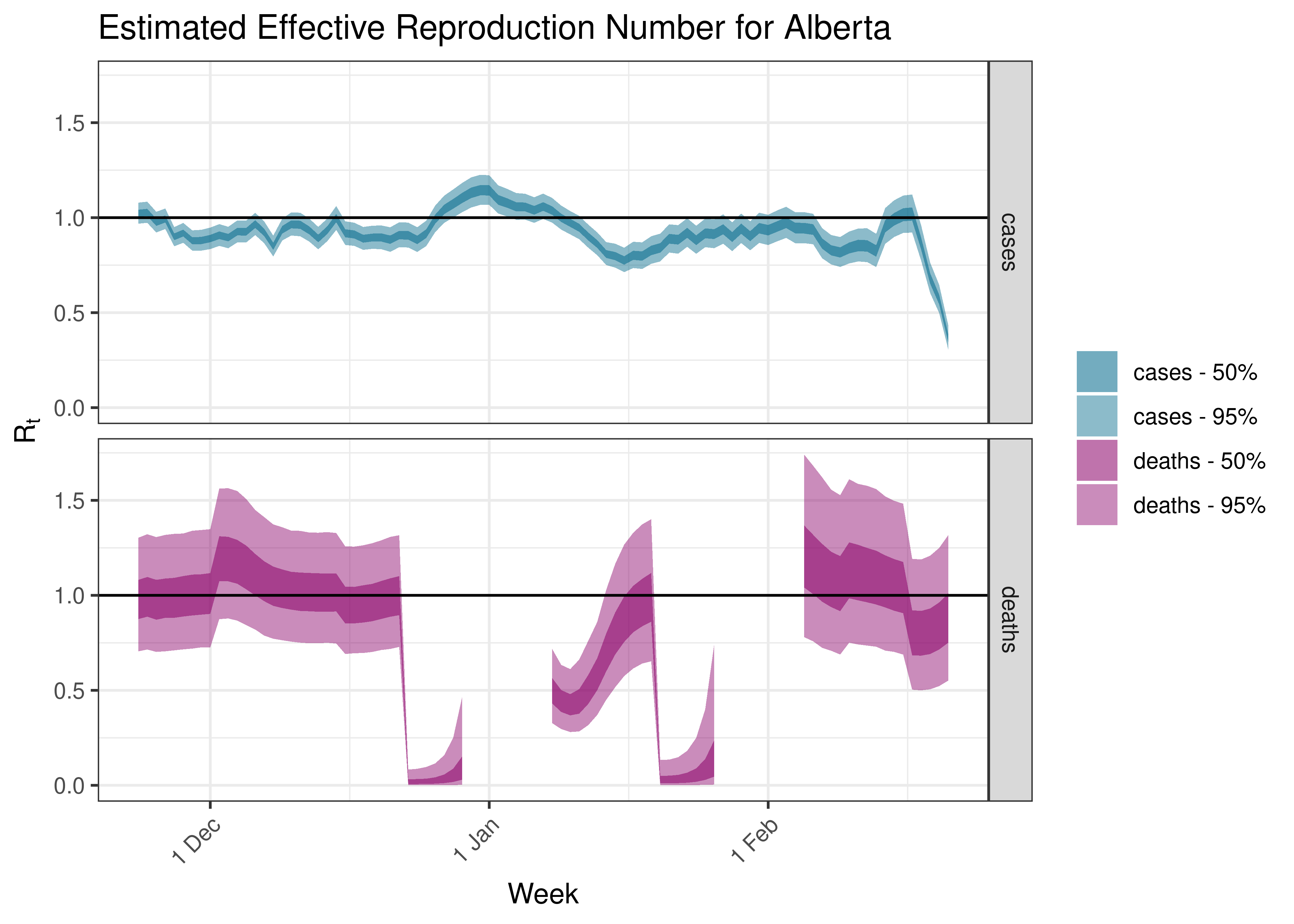
5.5.2 British Columbia
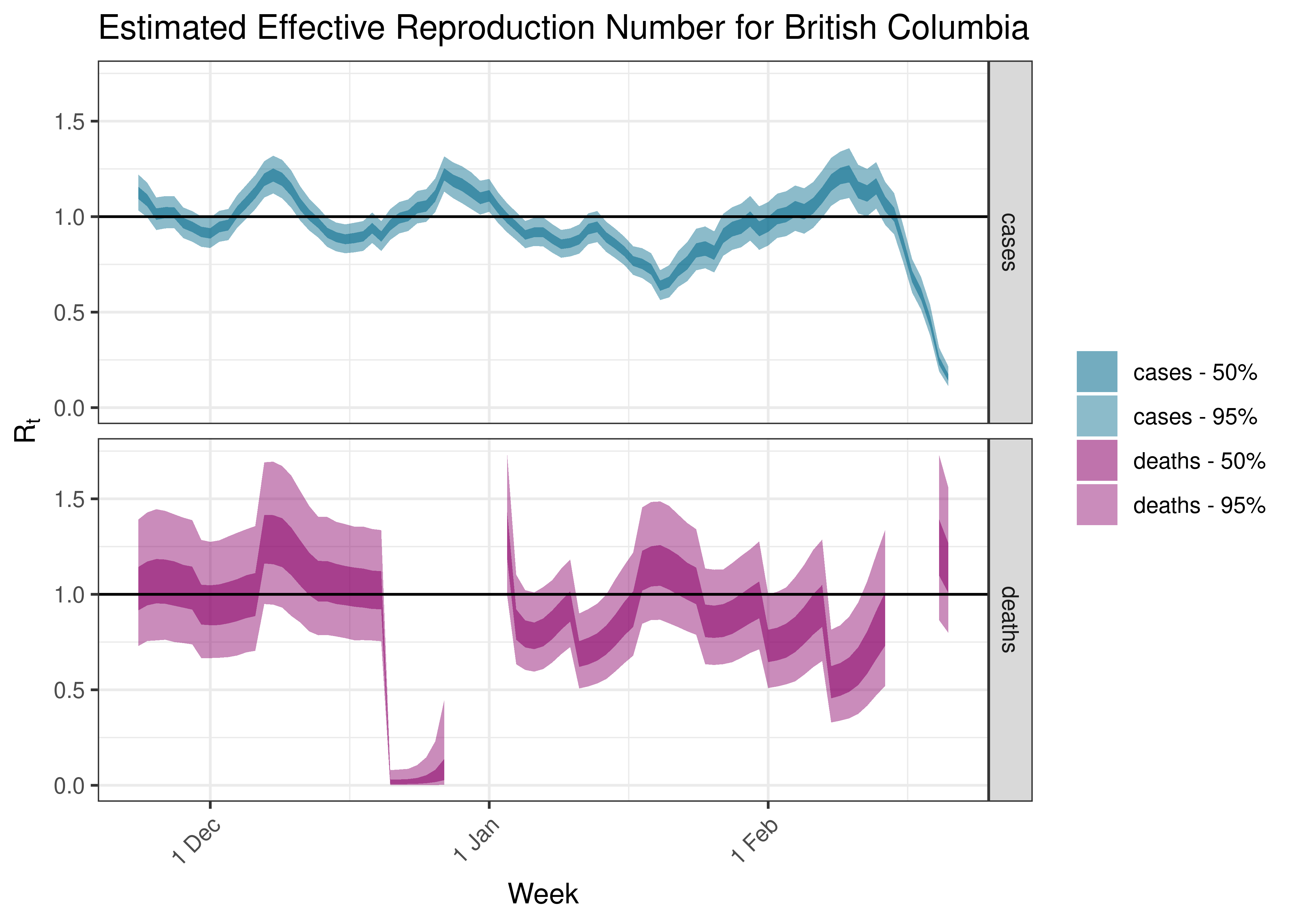
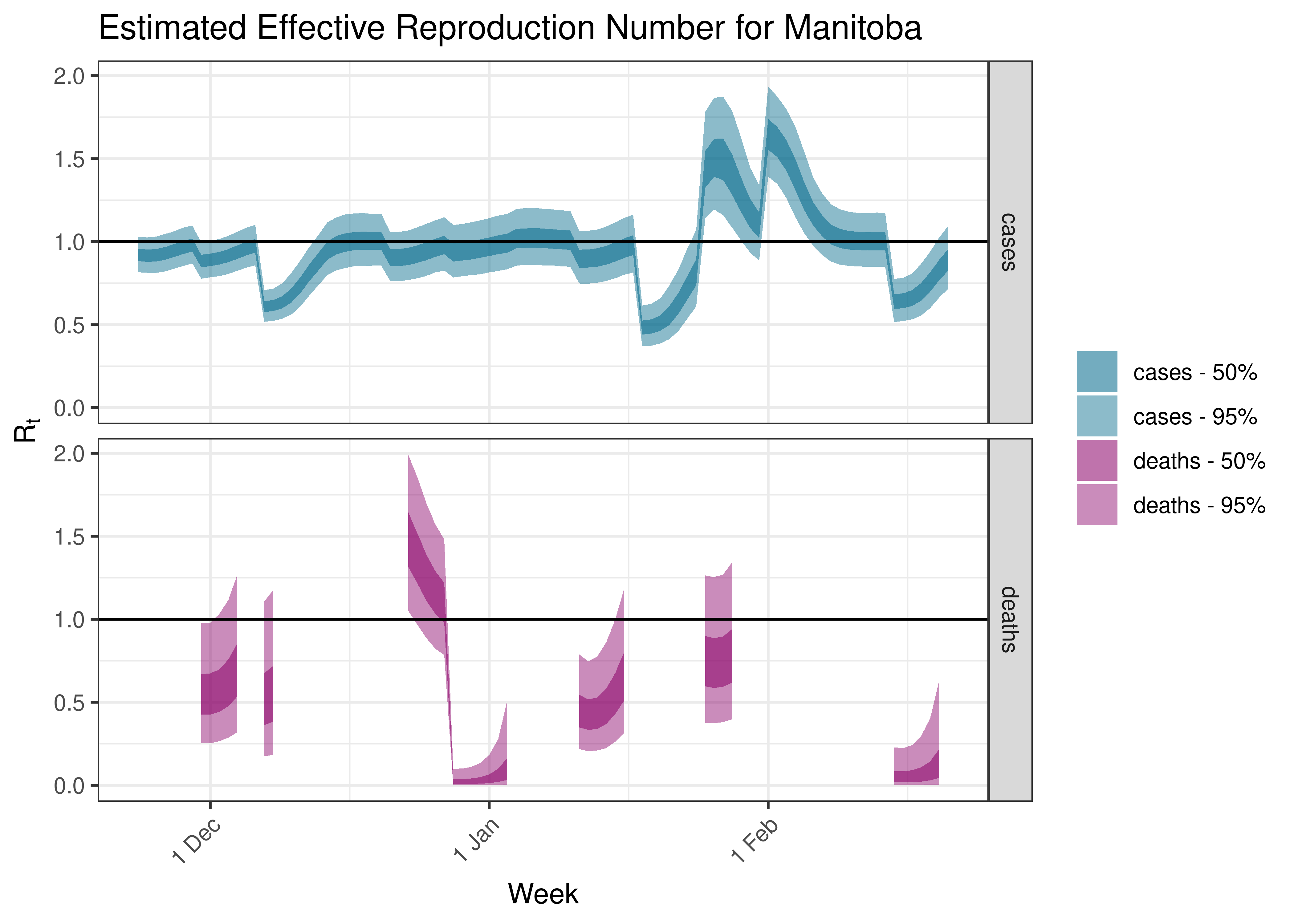
5.5.3 Manitoba
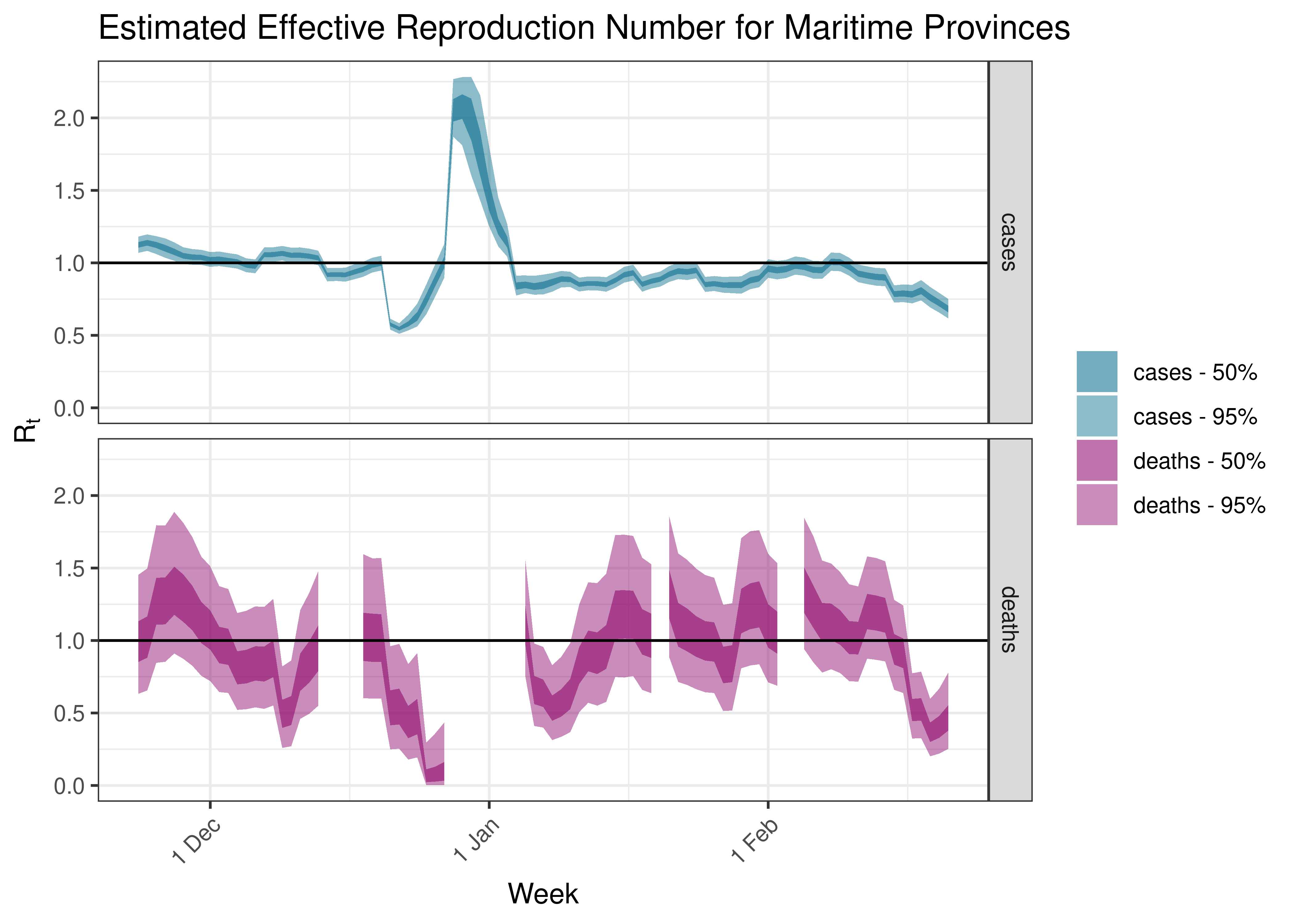
5.5.4 Maritime Provinces
5.5.5 Territories
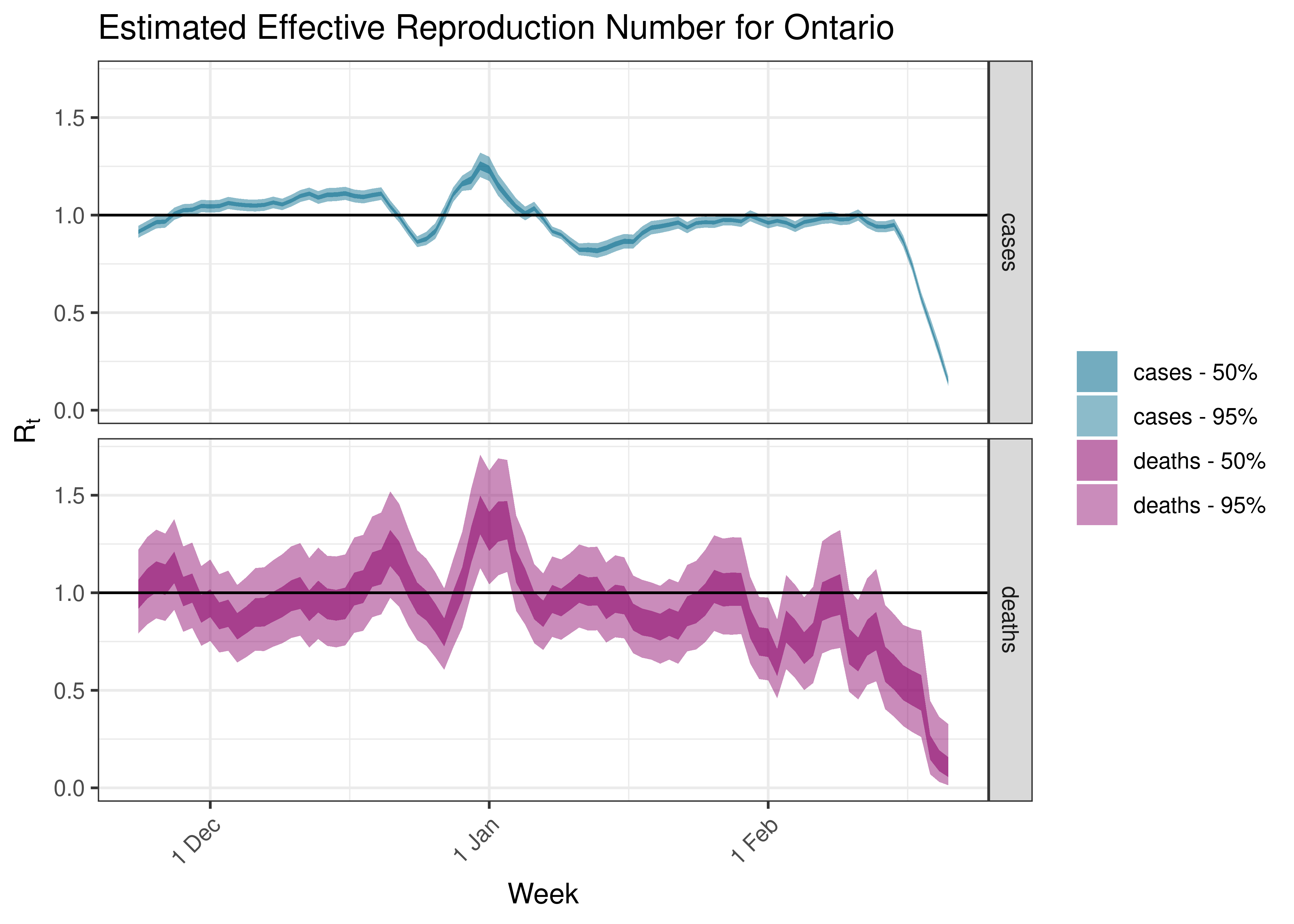
5.5.6 Ontario
5.5.7 Quebec
5.5.8 Saskatchewan
5.6 Detailed Results
Detailed output for all provinces are saved to a comma-separated value file. The file can be found here.
6 Discussion
Limitation of this method to estimate reproduction number are noted in [1]
- It’s sensitive to changes in transmissibility, changes in contact patterns, depletion of the susceptible population and control measures.
- It relies on an assumed generation interval assumptions.
- The size of the time window can affect the volatility of results.
- Results are time lagged with regards to true infection, more so in the case of the use of deaths.
- It’s sensitive to changes in case (or death) detection.
- The generation interval may change over time.
Further to the above the estimates are made under assumption that the cases and deaths are reported consistently over time. For cases this means that testing needs to be at similar levels and reported with similar lag. Should these change rapidly over an interval of a few weeks the above estimates of the effective reproduction numbers would be biased. For example a rapid expansion of testing over the last 3 weeks would results in overestimating recent effective reproduction numbers. Similarly any changes in reporting (over time and underreporting) of deaths would also bias estimates of the reproduction number estimated using deaths.
Estimates for the reproduction number are plotted in time period in which the relevant measure is recorded. Though in reality the infections giving rise to those estimates would have occurred roughly between a week to 4 weeks earlier depending on whether it was cases or deaths. These figures have not been shifted back.
Despite these limitation we believe the ease of calculation of this method and the ability to use multiple sources makes it useful as a monitoring tool.